PowerFit fits your 3D structures in any map!
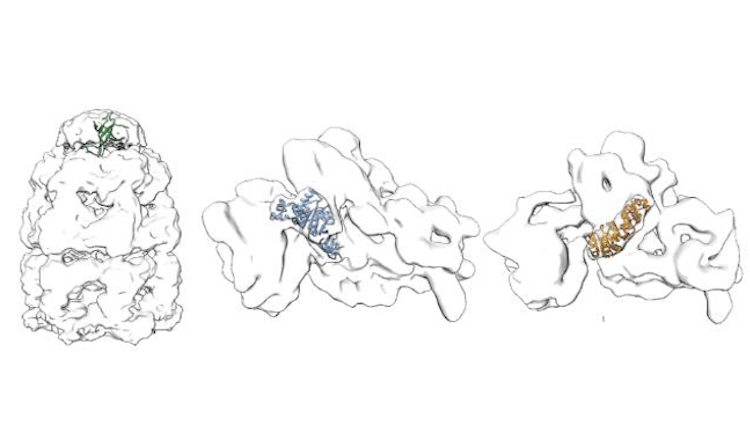
- PowerFit automatically fits high-resolution atomic structures into cryo-EM densities.
To this end it performs a full-exhaustive 6-dimensional cross-correlation search between the atomic structure and the density. It takes as input an atomic structure in PDB- or mmCIF-format and a cryo-EM density with its resolution; and outputs positions and rotations of the atomic structure corresponding to high correlation values. PowerFit uses the local cross-correlation function as its base score. The score is by default enhanced with an optional Laplace pre-filter and a core-weighted version to minimize overlapping densities from neighboring subunits. - In order to constantly improve our service and give you the best experience, we would really appreciate if you could take the time to complete our short online survey (~5min) available here: https://goo.gl/forms/mmO4PLP8SCweKr5r2
- PowerFit webserver
- REGISTRATION: To use the PowerFit server you must have registered for an account. If you do not have an account yet you can register here
- Submit your job to:
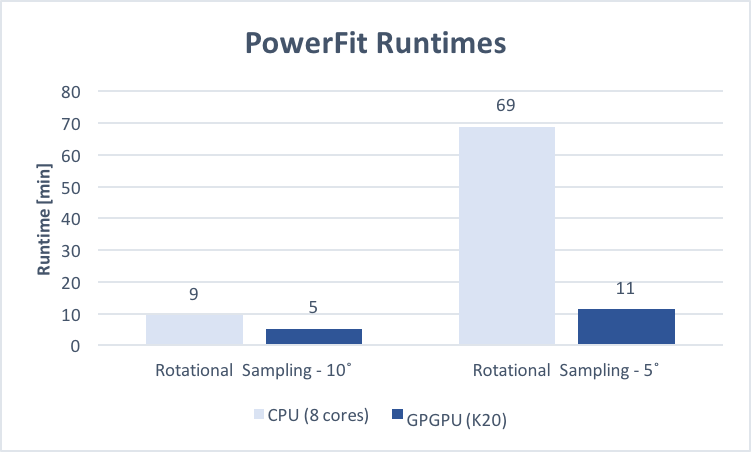
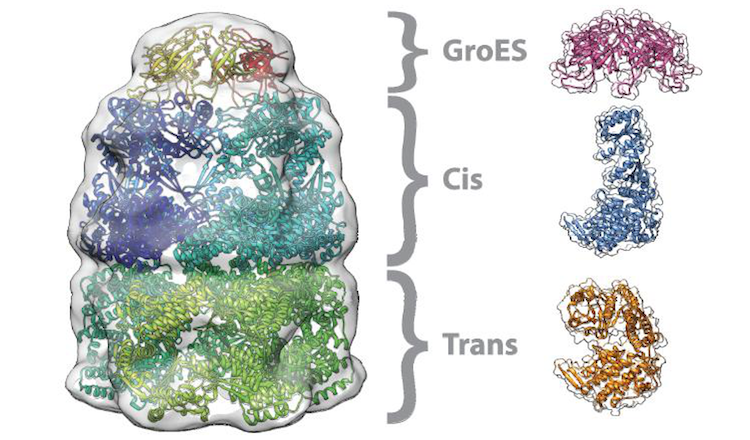
Average run time in minutes (5 runs each, excluding pre- and post-processing, and queued time) for fitting
chain C of the GroEL-ES complex (PDB: 3zpz, 525 residues)
into the corresponding map (EMD-2325,
Resolution: 8.9 Å, Estimated Volume: 936 nm3) with a rotational sampling
interval of 5° and 10°, submitted to the local CPU and grid GPGPU resources via the PowerFit web server.
- Powerfit Webinar
-
Watch the 11th BioExcel Webinar on “Robust solutions for cryoEM fitting and visualisation of interaction space”. This Webinar presented by the PowerFit team features a comprehensive overview about the general methodology and algorithms behind PowerFit. Furthermore it provides a quick introduction to the PowerFit web portal implementation including a live demo of the portal.
- Reference for use of the server
- When using the PowerFit server please cite:
- R. Vargas Honorato, P.I. Koukos, B. Jimenez-Garcia, A. Tsaregorodtsev, M. Verlato, A. Giachetti, A. Rosato and A.M.J.J. Bonvin.
Structural biology in the clouds: The WeNMR-EOSC Ecosystem. Frontiers Mol. Biosci., 8, fmolb.2021.729513 (2021).
G.C.P. van Zundert, M. Trellet, J. Schaarschmidt, Z. Kurkcuoglu, M. David, M. Verlato, A. Rosato and A.M.J.J. Bonvin.
The DisVis and PowerFit web servers: Explorative and Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 429, 399-407 (2017).
G.C.P. van Zundert and A.M.J.J. Bonvin (2015)
Fast and sensitive rigid-body fitting into cryo-EM density maps with PowerFit. AIMS Biophysics 2, 73-87.
and add the following acknowledgment:
The FP7 WeNMR (project# 261572), H2020 West-Life (project# 675858), EOSC-hub (project# 777536) and the EGI-ACE (project# 101017567) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure with the dedicated support of CESNET-MCC, INFN-LNL-2, NCG-INGRID-PT, TW-NCHC, CESGA, IFCA-LCG2, UA-BITP, TR-FC1-ULAKBIM, CSTCLOUD-EGI, IN2P3-CPPM, CIRMMP, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, Taiwan and the US Open Science Grid.- PowerFit software
- You can also install PowerFit directly on your computer. Source code is available under Apache 2.0 license here: Github::PowerFit