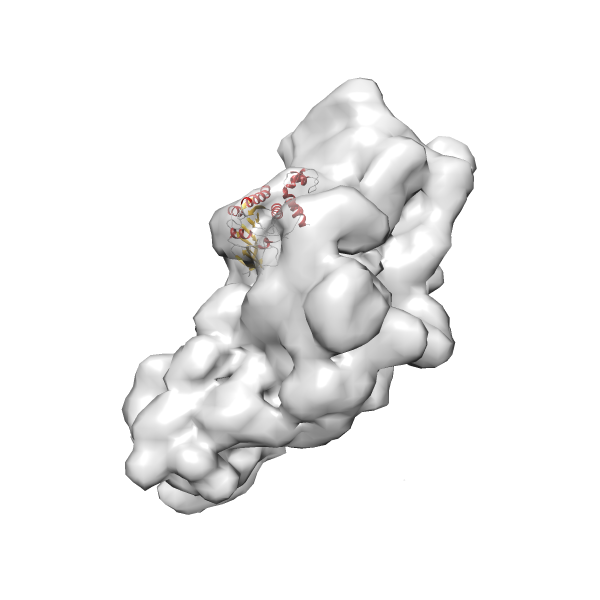
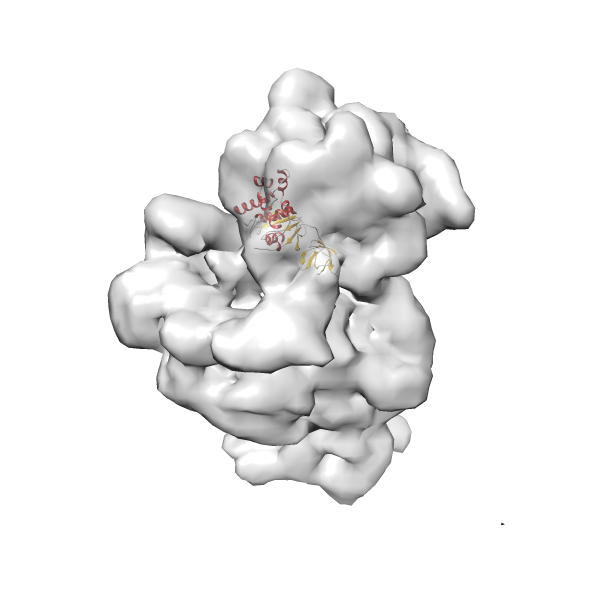
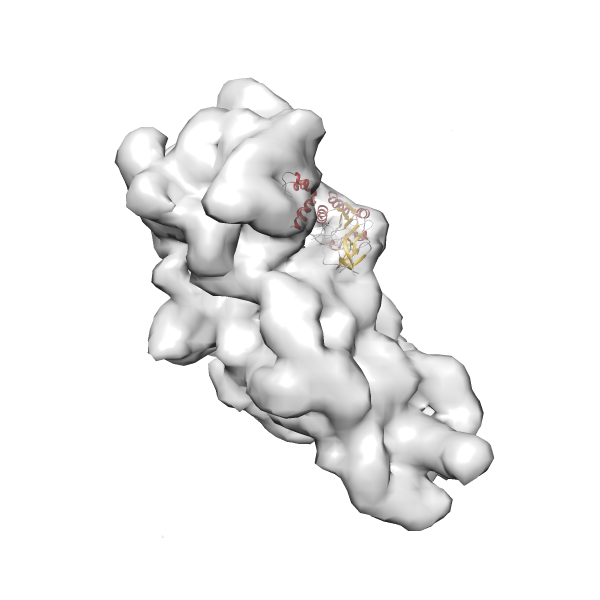
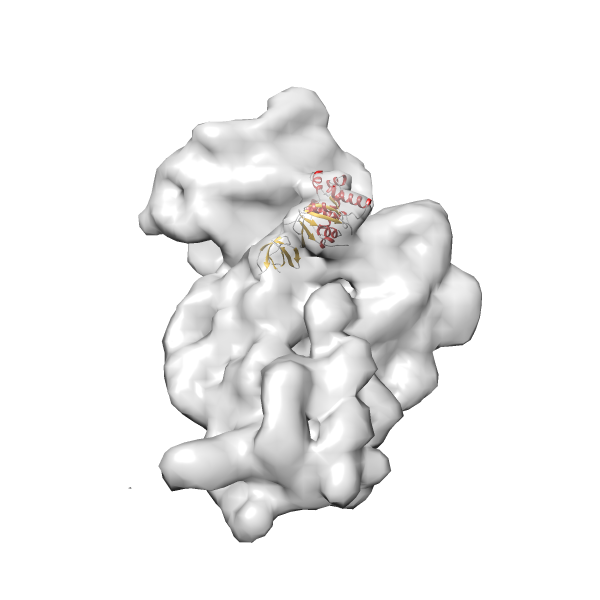
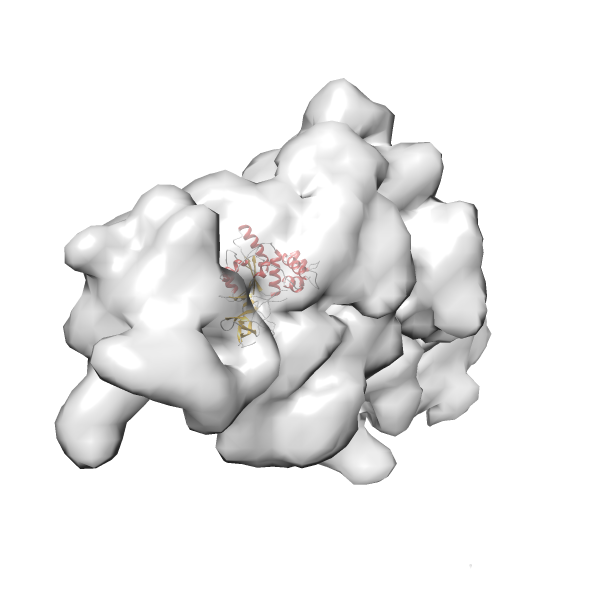
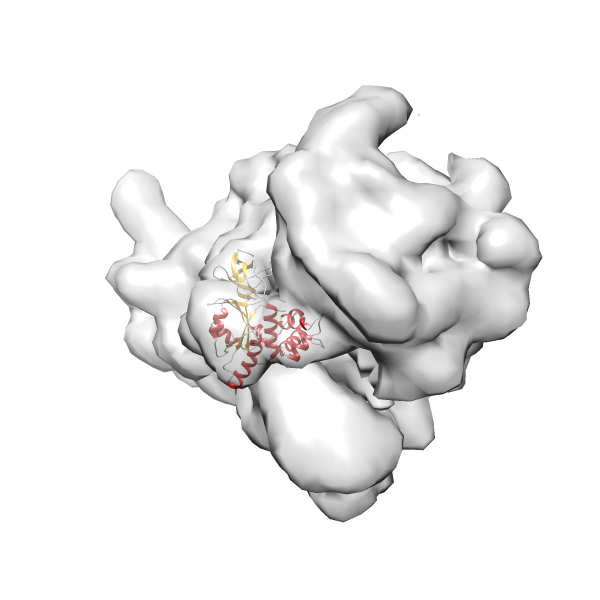
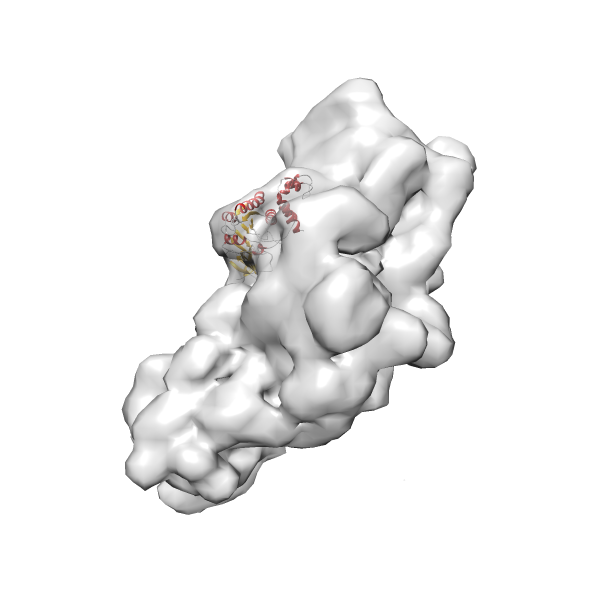
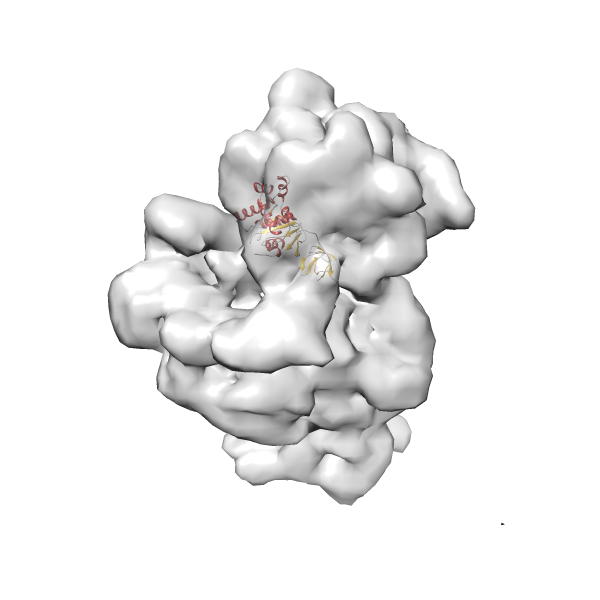
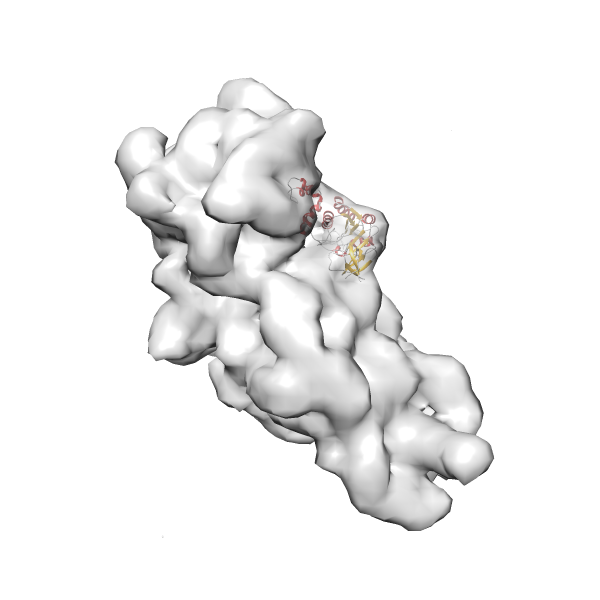
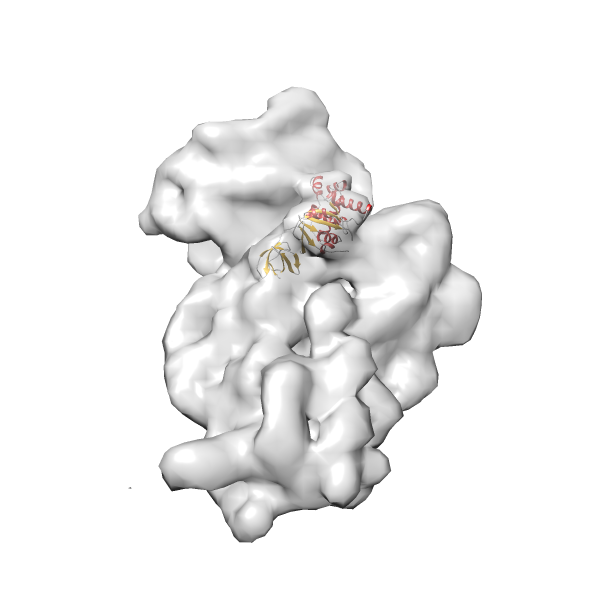
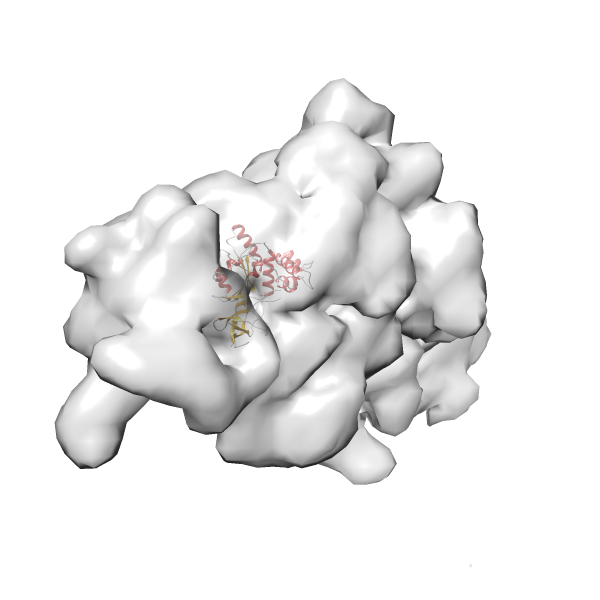
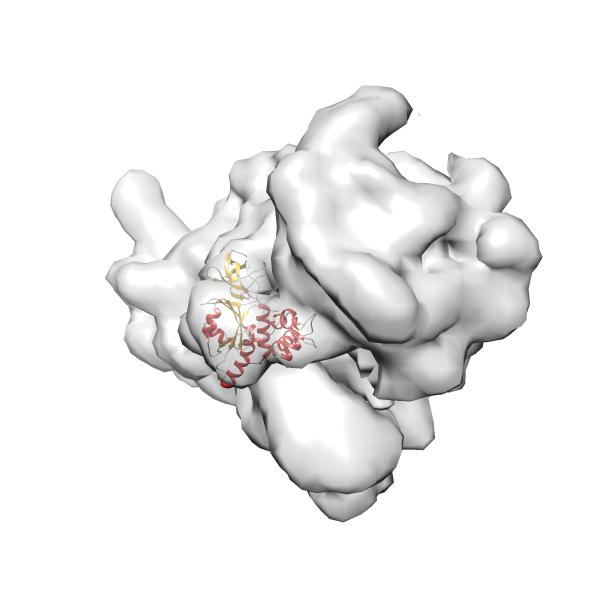
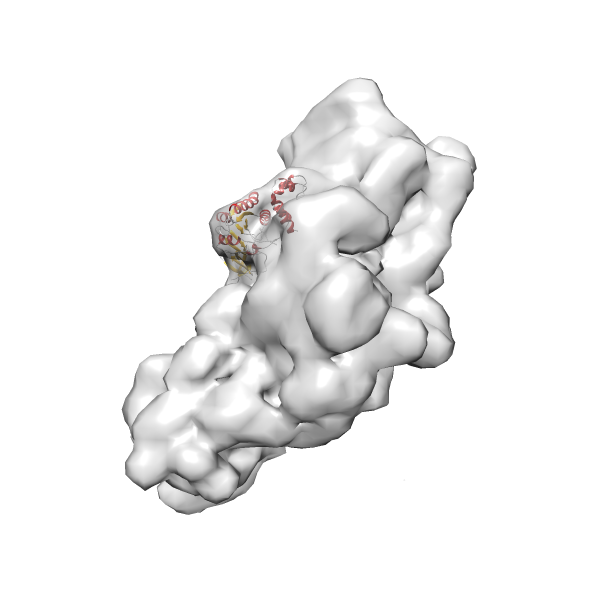
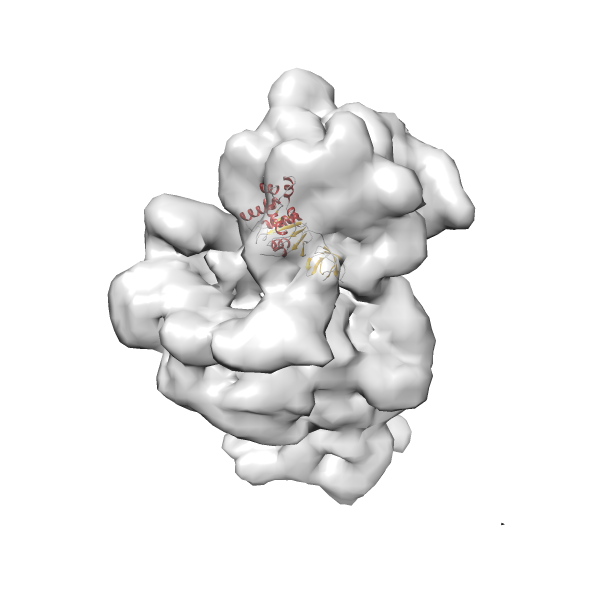
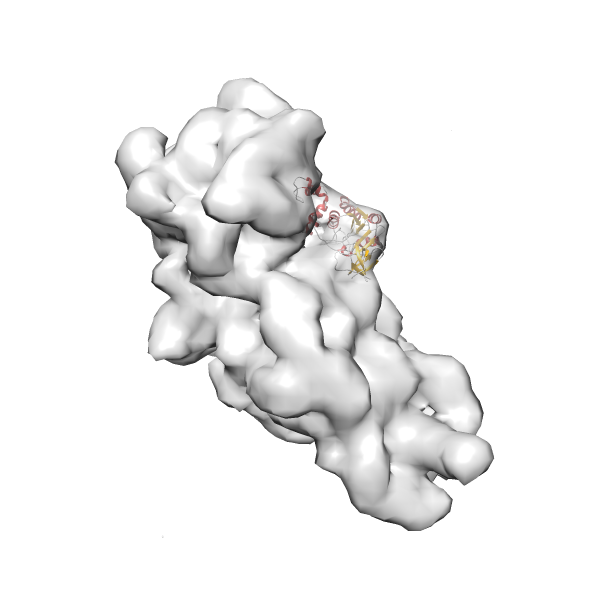
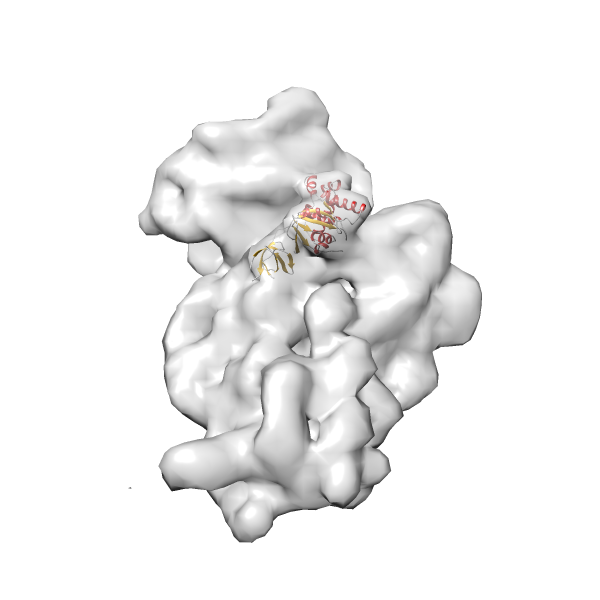
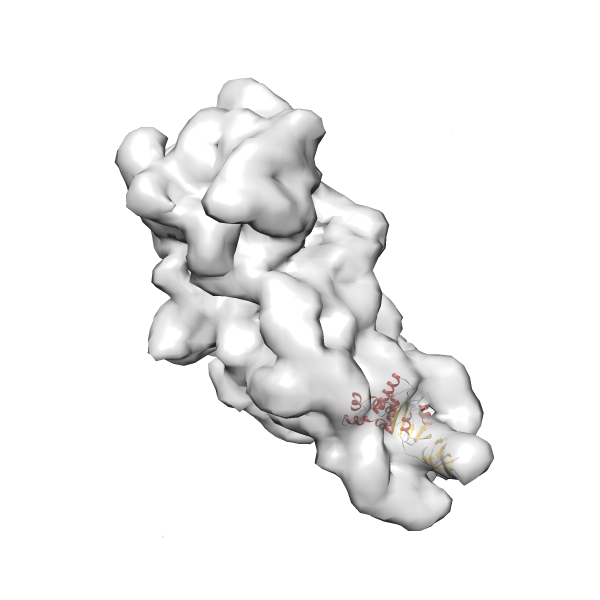
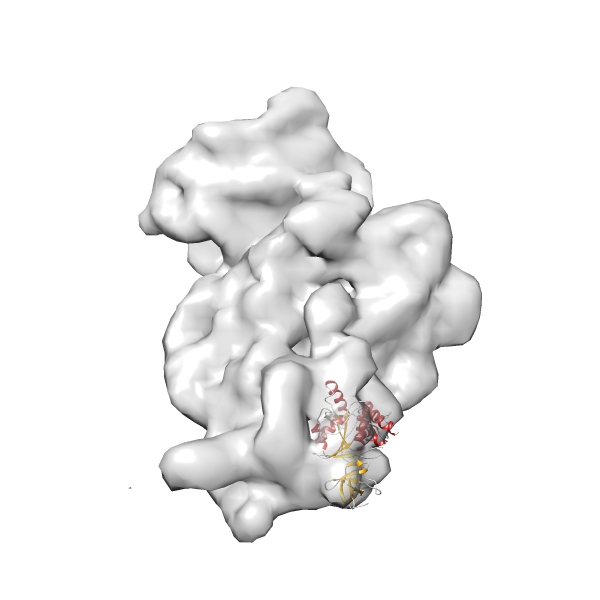
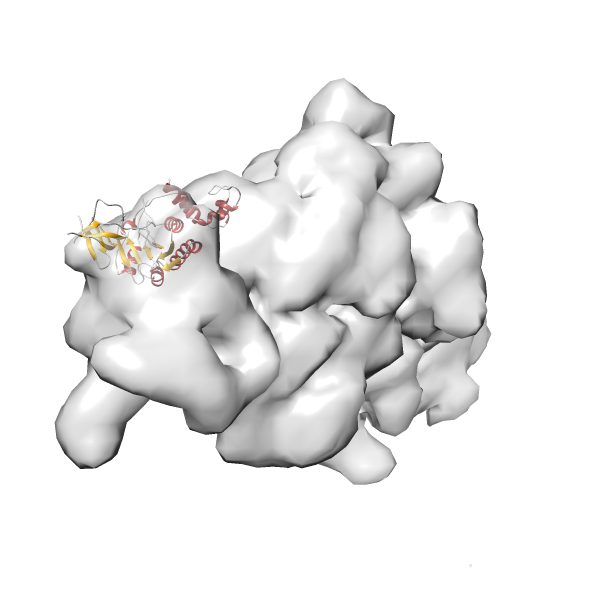
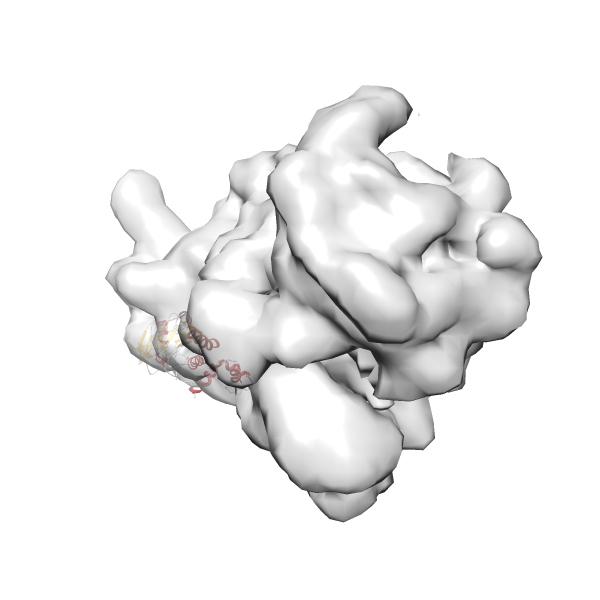
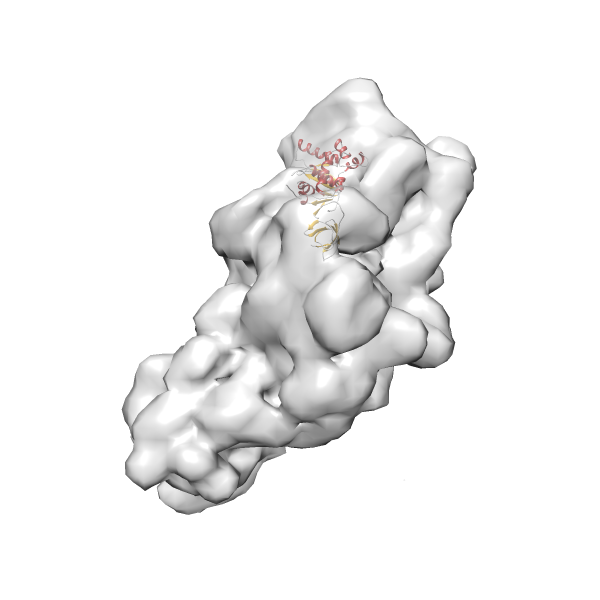
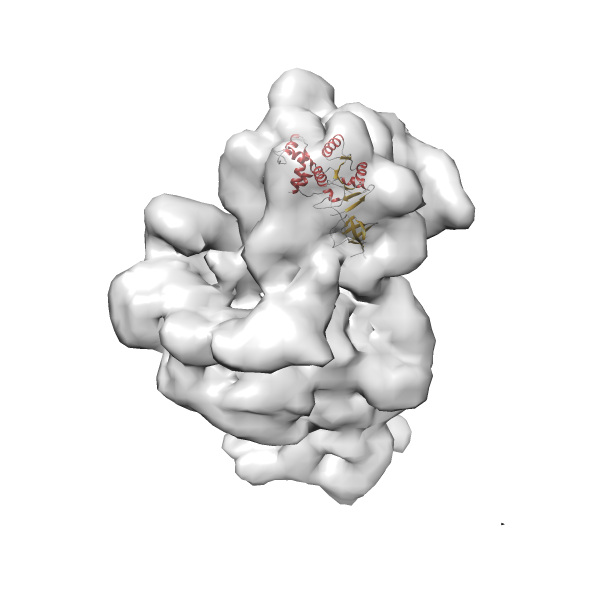
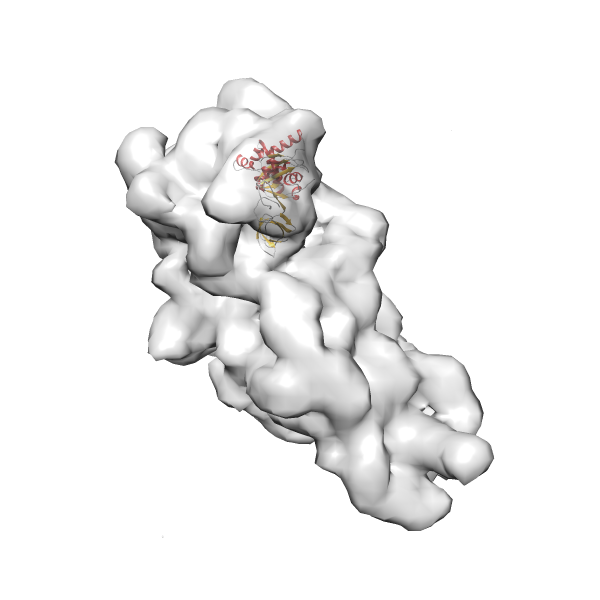
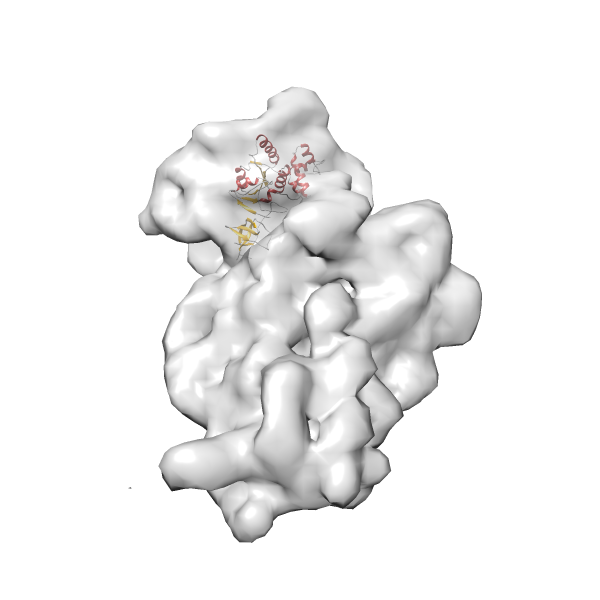
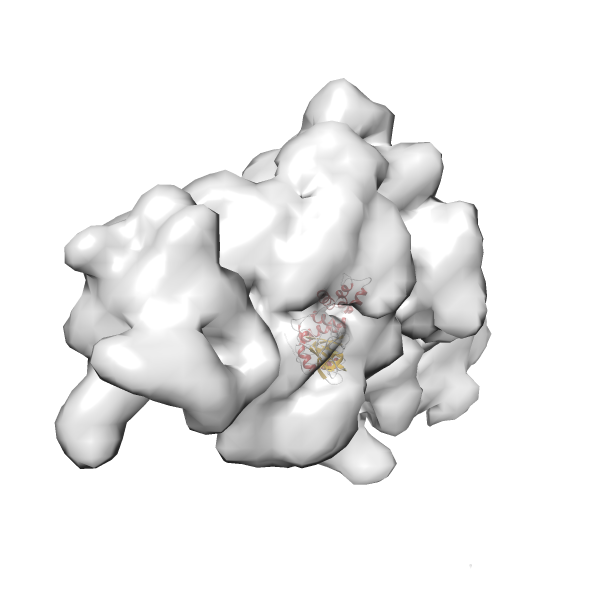
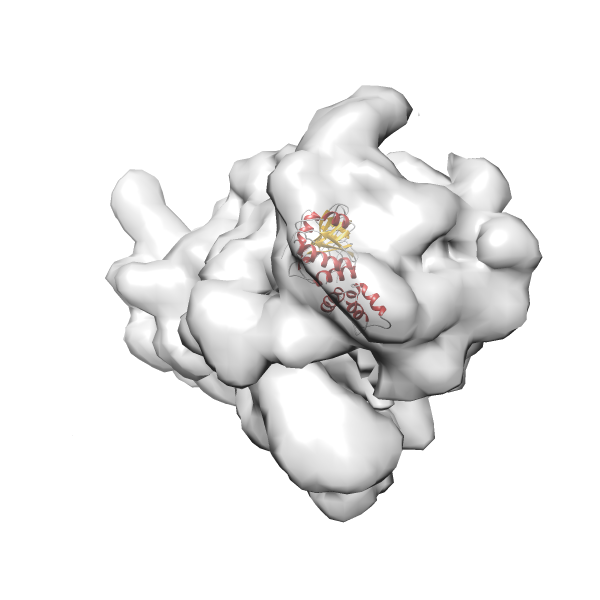
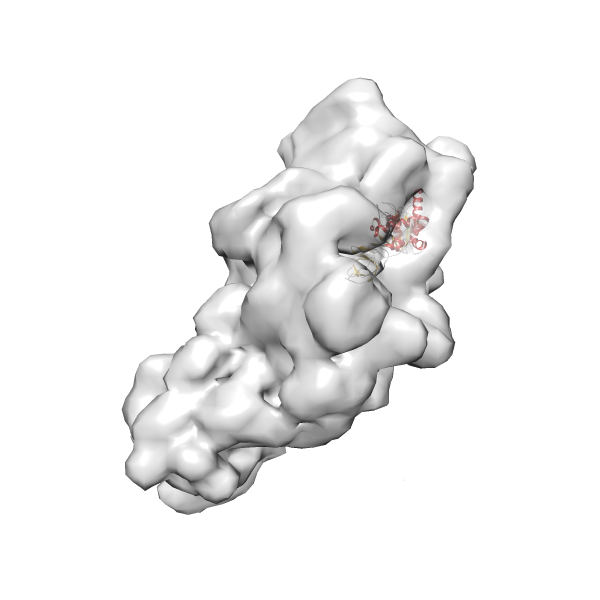
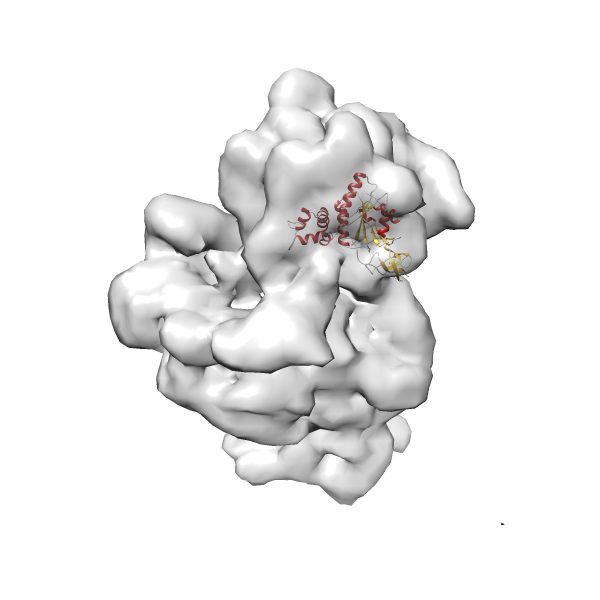
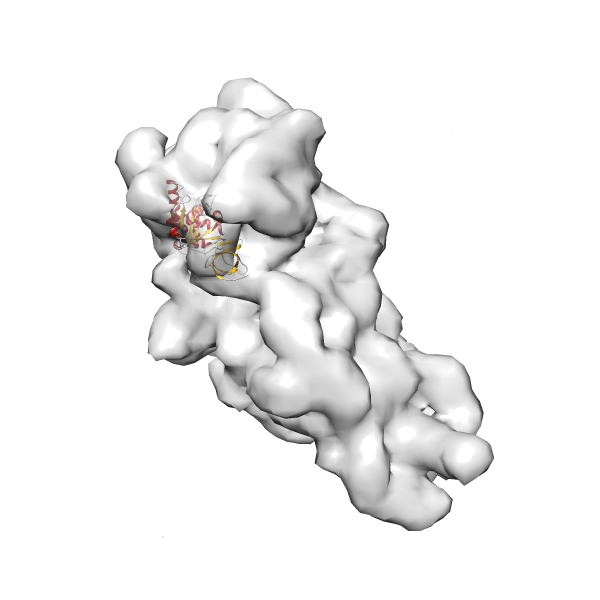
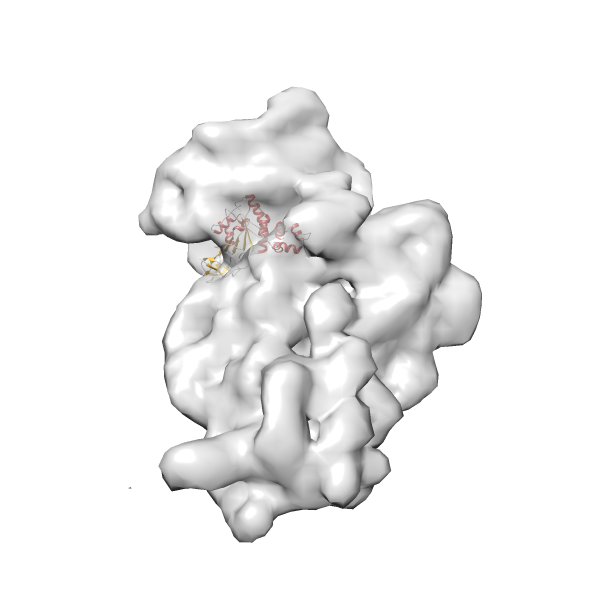
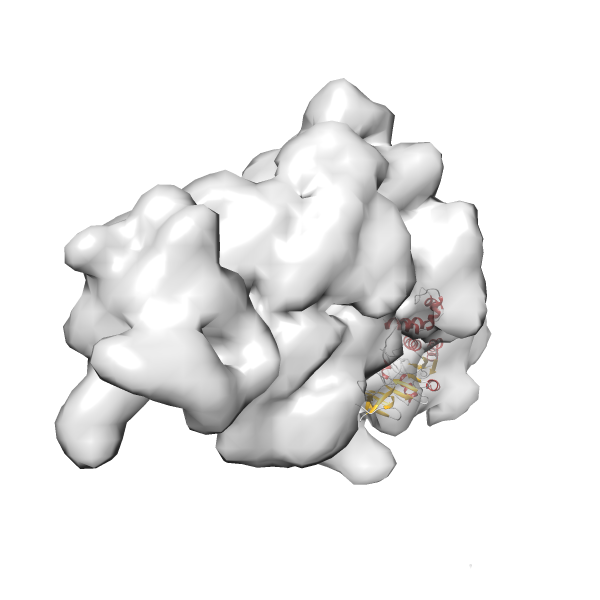
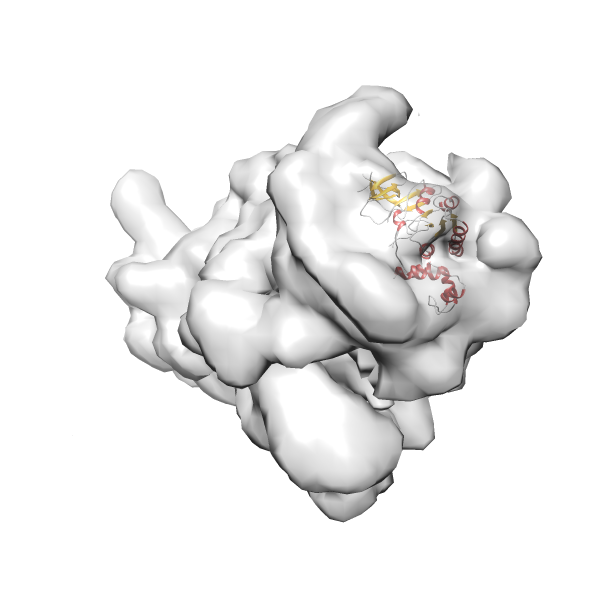
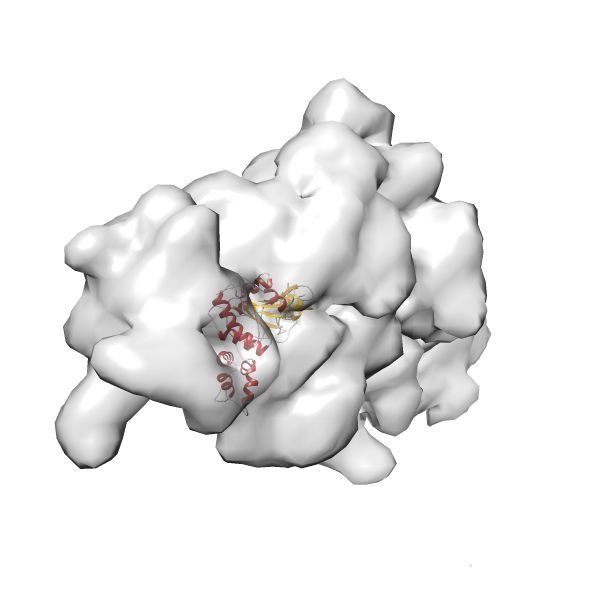
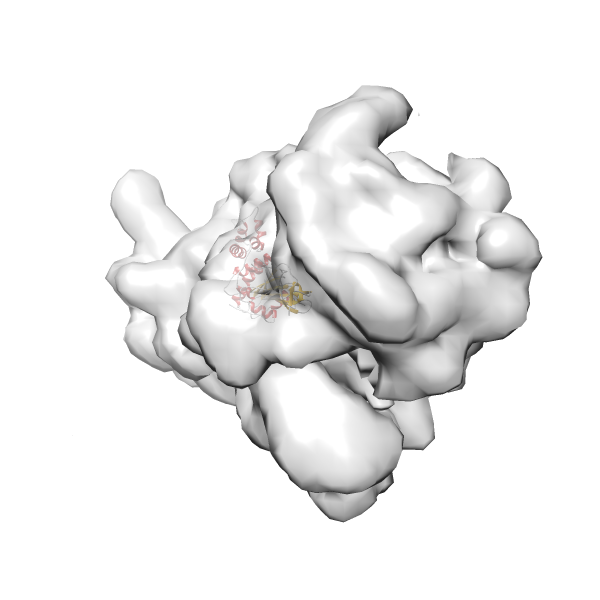
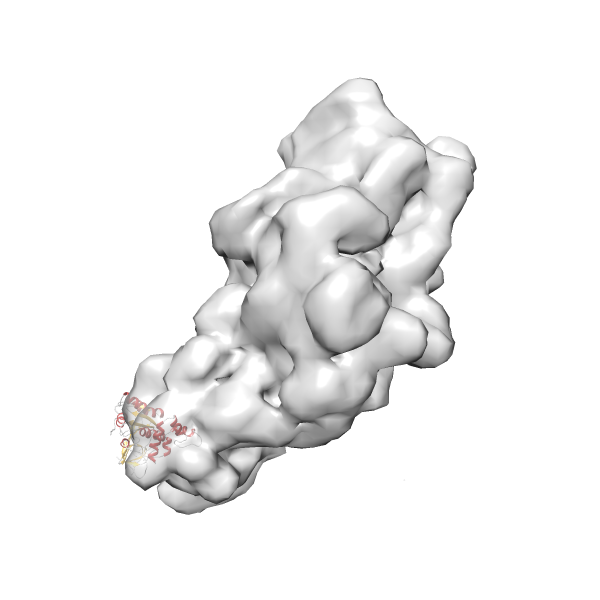
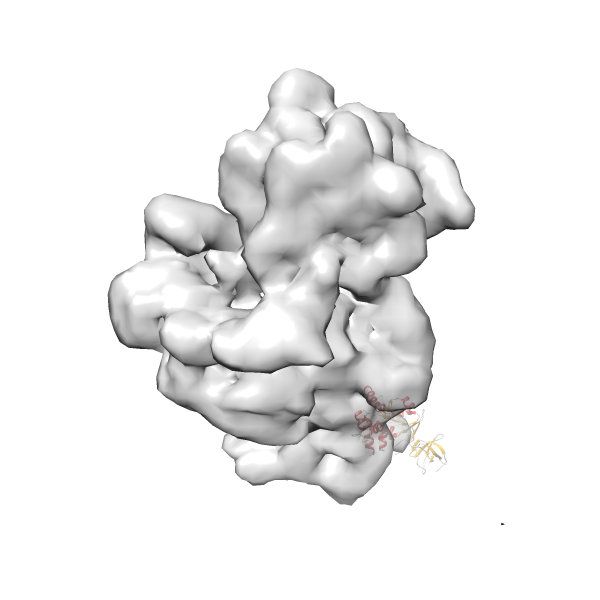
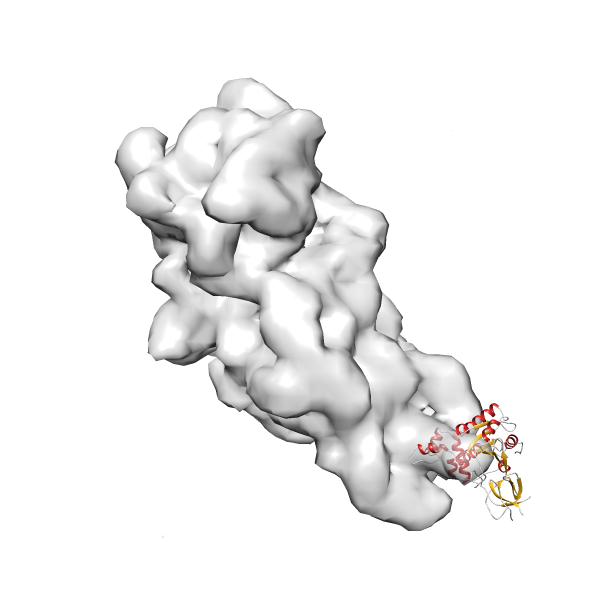
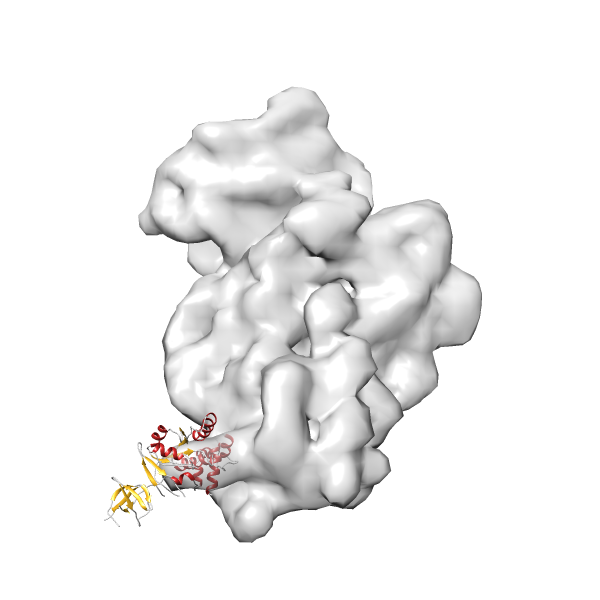
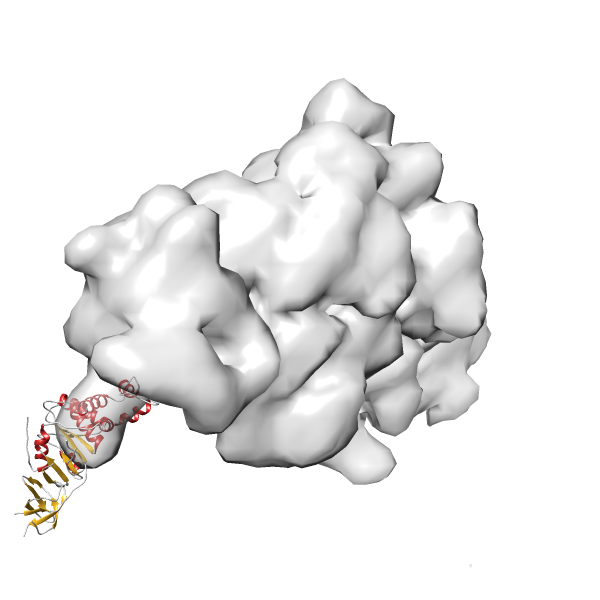
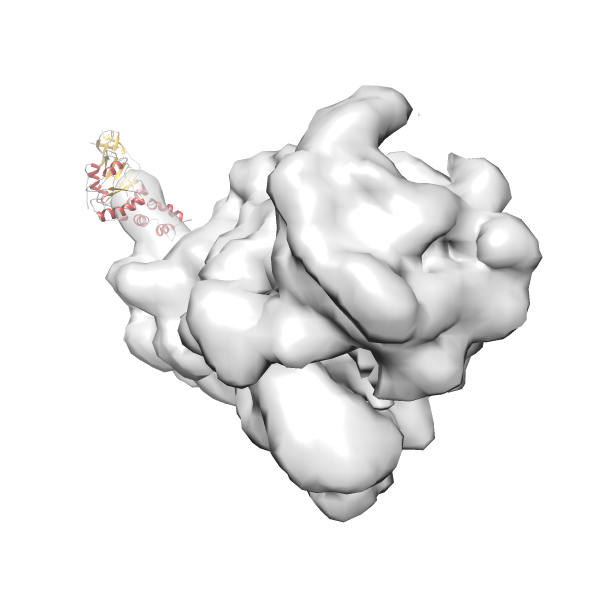
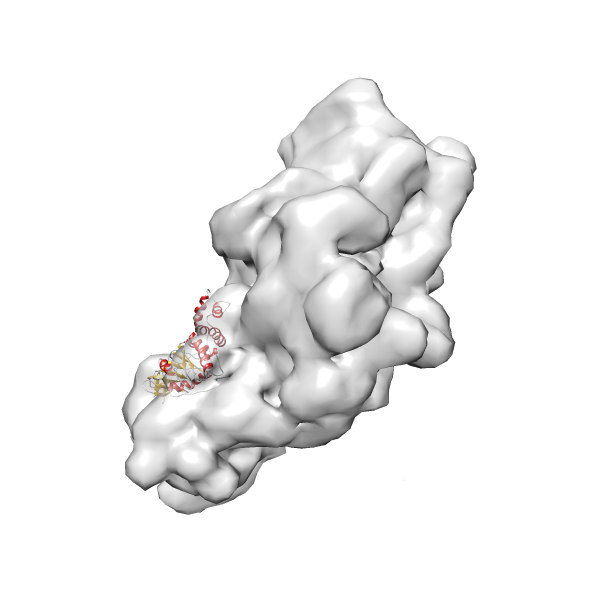
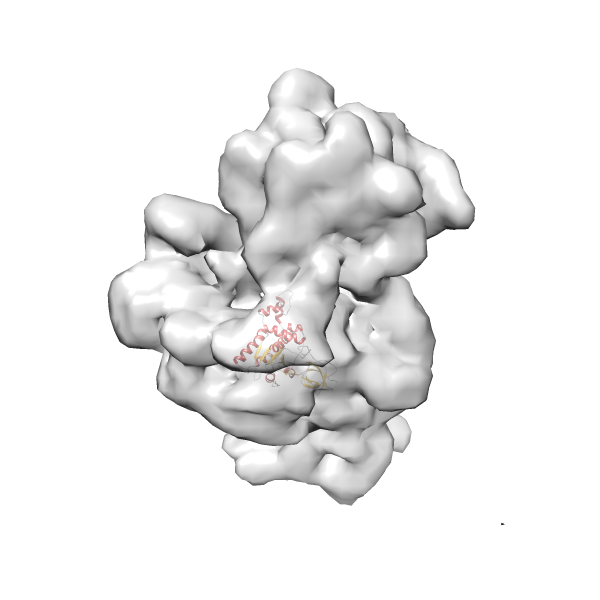
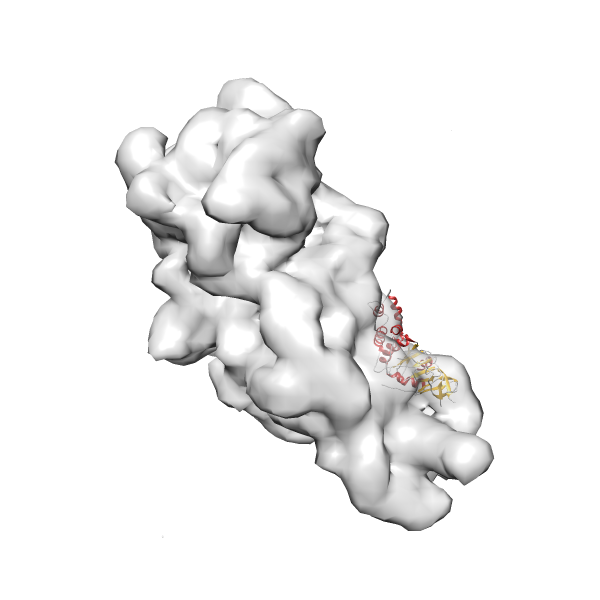
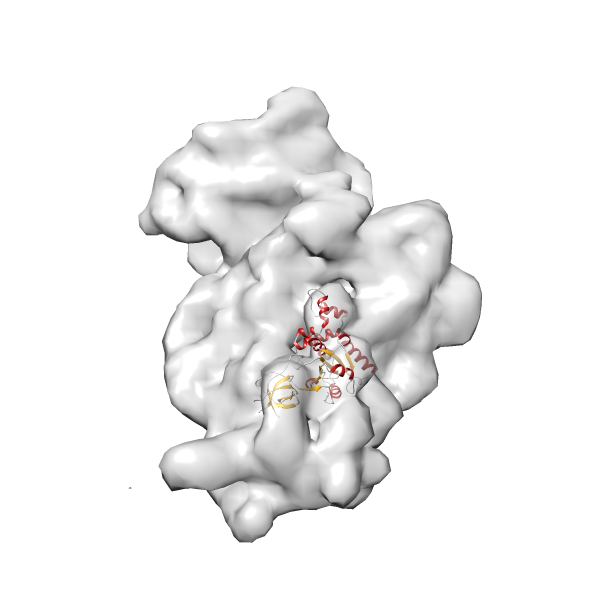
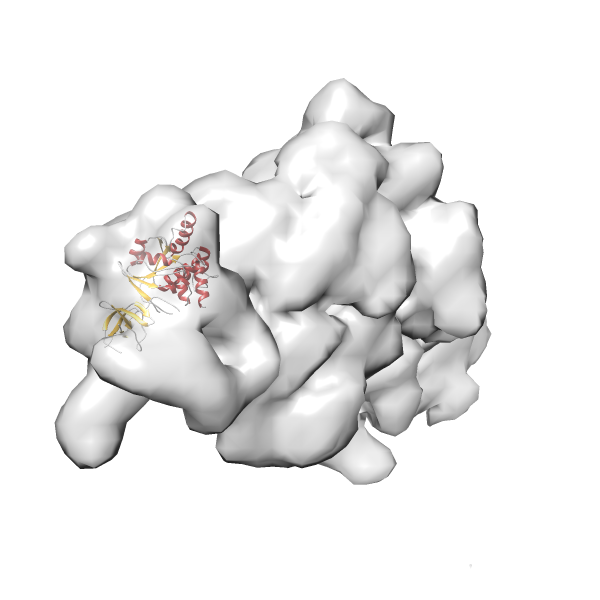
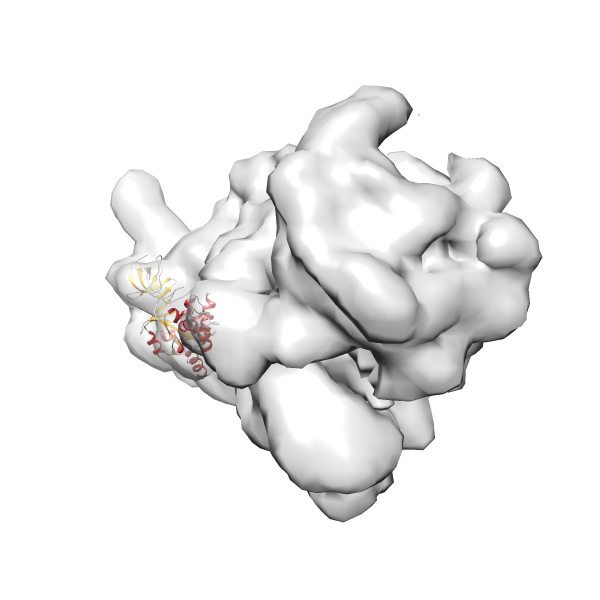
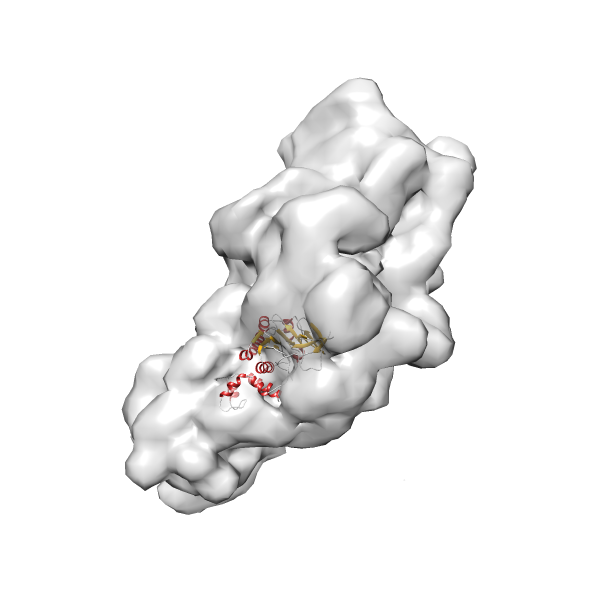
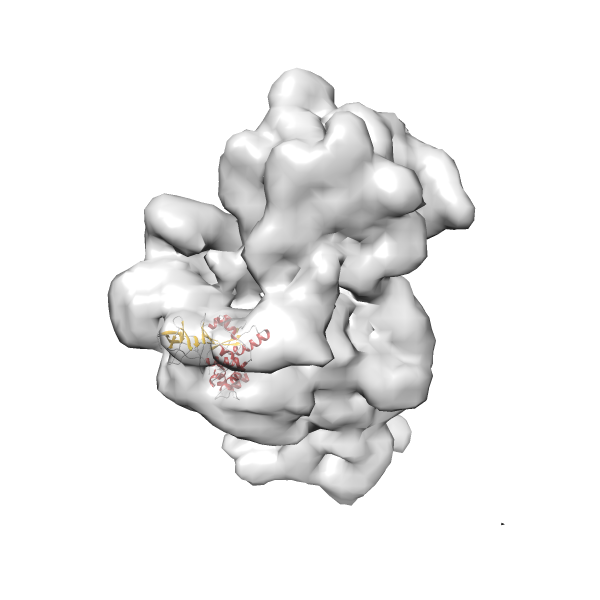
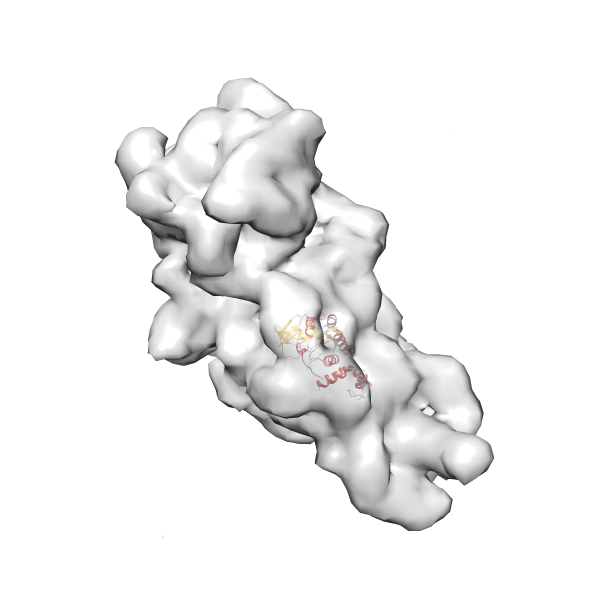
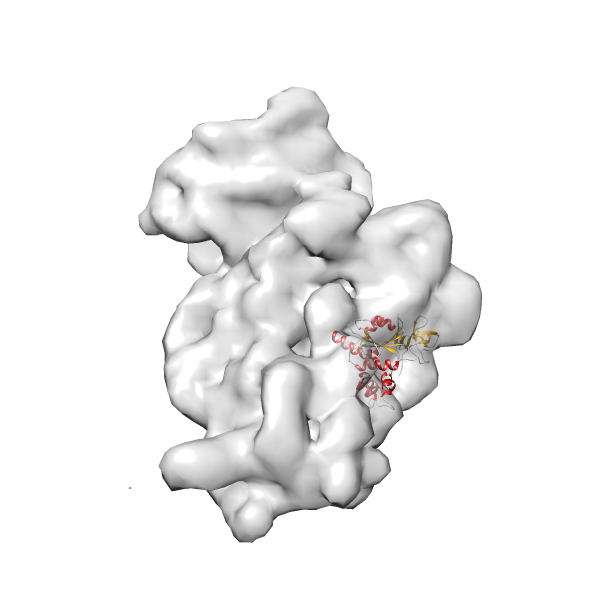
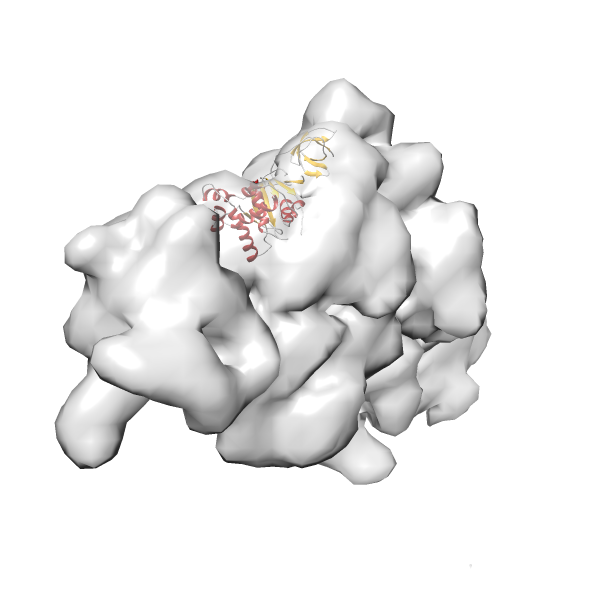
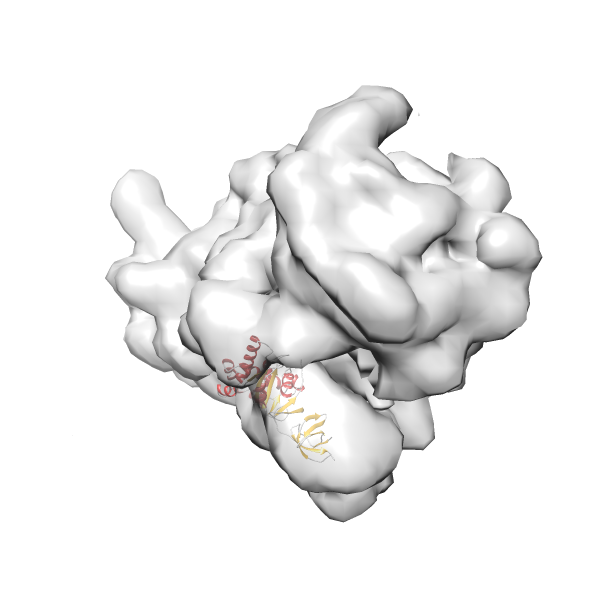
- GTPase RsgA-30S ribosomal subunit-GMPPNP complex
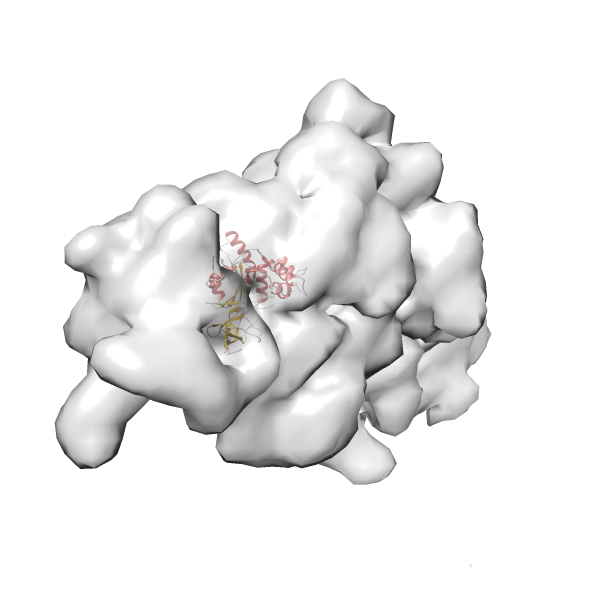
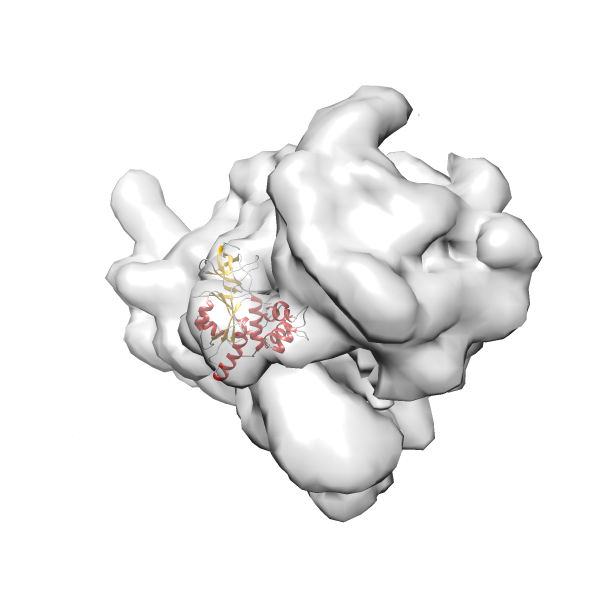
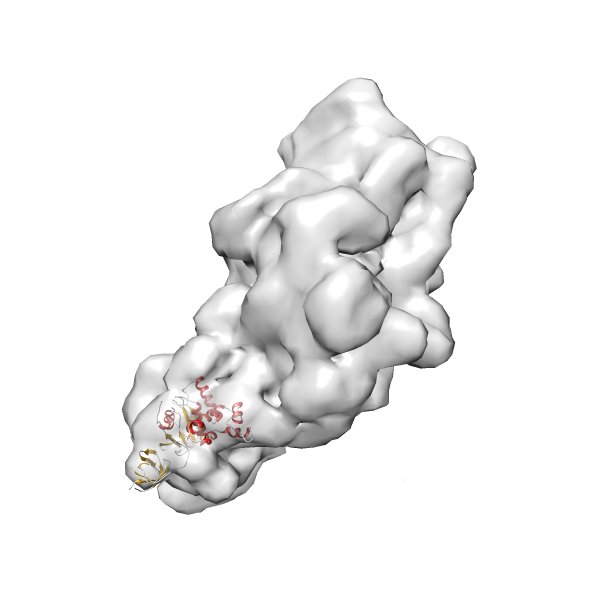
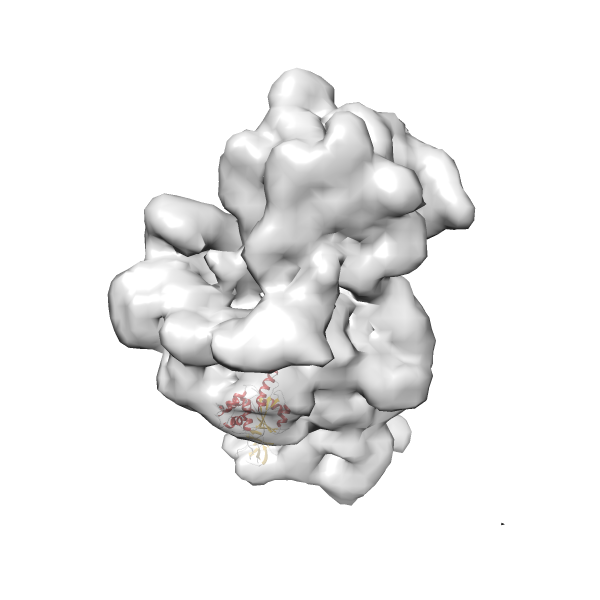
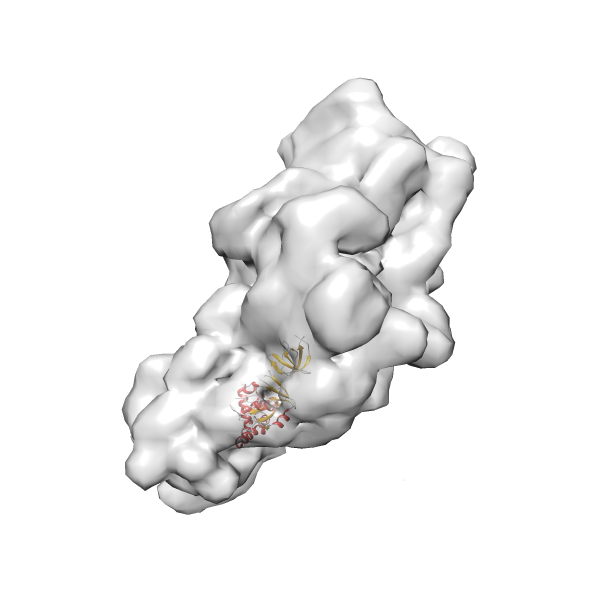
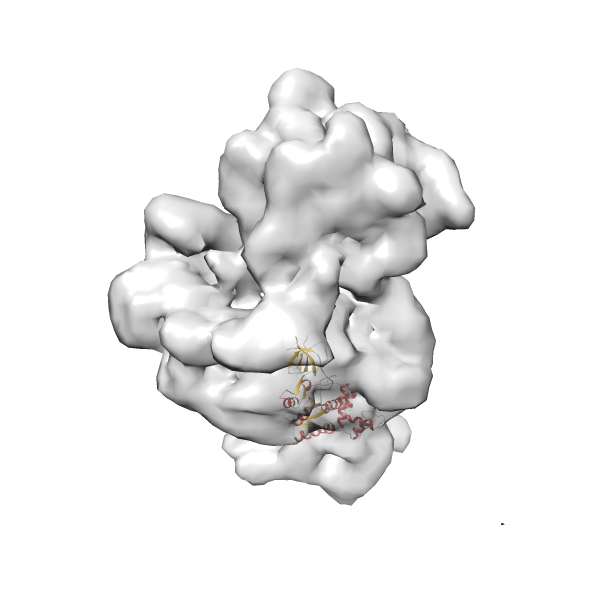
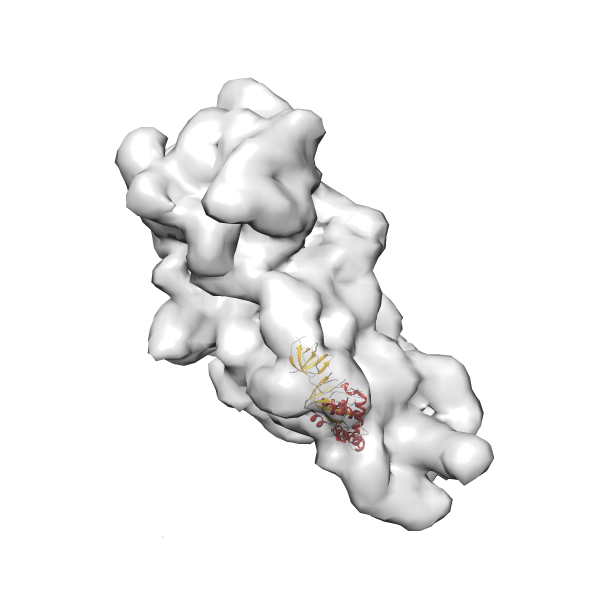
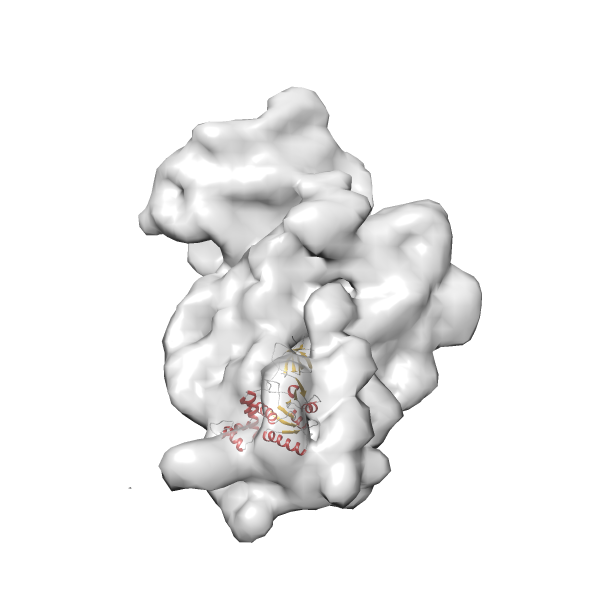
- In this example, which is also used within the scope of the PowerFit web server tutorial, the putative ribosome biogenesis GTPase RsgA (chain W of PDB entry 2ykr) was fitted into the cryo-EM density map of the full RsgA-30S ribosomal subunit-GMPPNP complex (EMDB entry 1884 - 9.8 Å resolution) with the default server parameters. The best 3 Fits all occupy the expected part of the density map with powerfit clearly favoring one particular orientation based on cross correlation score and sigma difference.
- Run 2ykr_W - EMD-1884 (Example1)
- Status: FINISHED
- Your PowerFit run has successfully completed.
-
Archive of the complete run: Example1.tgz Archive of all autogenerated images: Example1_images.tgz - Please cite the following papers in your work:
-
G.C.P. van Zundert, M. Trellet, J. Schaarschmidt, Z. Kurkcuoglu, M. David, M. Verlato, A. Rosato and A.M.J.J. Bonvin.
The DisVis and PowerFit web servers: Explorative and Integrative Modeling of Biomolecular Complexes. J. Mol. Biol., Advanced Online Publication (2016).
G.C.P. van Zundert and A.M.J.J. Bonvin (2015)
Fast and sensitive rigid-body fitting into cryo-EM density maps with PowerFit.
AIMS Biophysics 2, 73-87. - and add the following acknowledgment:
- The FP7 WeNMR (project# 261572), H2020 West-Life (project# 675858), EOSC-hub (project# 777536) and the EGI-ACE (project# 101017567) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure with the dedicated support of CESNET-MCC, INFN-LNL-2, NCG-INGRID-PT, TW-NCHC, CESGA, IFCA-LCG2, UA-BITP, TR-FC1-ULAKBIM, CSTCLOUD-EGI, IN2P3-CPPM, CIRMMP, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, Taiwan and the US Open Science Grid.
- Note: Your results will be stored for 14 days before being removed from the server, please make a backup as soon as possible.
- How would you rate your experience with our portal? sentiment_very_dissatisfied sentiment_dissatisfied sentiment_neutral sentiment_satisfied sentiment_very_satisfied Thank you!done
- Questions / feedback ? ask.bioexcel.eu
- Please also consider giving us some feedback by filling our online survey.
- Solutions
- The table below lists the 15 best non-redundant solutions found by correlation score. The first column shows the rank, column 2 the correlation score, column 3 the Fisher z-score column 4 the zscore as factor of standard deviations (z/σ), and column 5 the sigma difference to the best fit. (see N. Volkmann 2009, and Van Zundert and Bonvin 2016).
-
Rank (N) Cross Correlation Score Fisher z-score z-score/σ Sigma difference
(z1-zN)/σ1 0.560 0.633 31.0 0.00 2 0.517 0.573 28.0 2.98 3 0.432 0.463 22.7 8.35 4 0.301 0.311 15.2 15.79 5 0.298 0.307 15.1 15.97 6 0.298 0.307 15.0 15.99 7 0.293 0.302 14.8 16.24 8 0.290 0.298 14.6 16.42 9 0.288 0.297 14.5 16.49 10 0.288 0.296 14.5 16.53 11 0.287 0.295 14.4 16.58 12 0.285 0.293 14.3 16.69 13 0.284 0.292 14.3 16.71 14 0.284 0.292 14.3 16.73 15 0.282 0.290 14.2 16.82 - Images were generated with UCSF Chimera.
- Fit 1
-
Rank 1 Cross Correlation Score 0.560 Fisher z-score 0.633 z-score/σ 31.0 Sigma difference (zN-zN+1)/σ 0.00 PDB Download - Fit 2
-
Rank 2 Cross Correlation Score 0.517 Fisher z-score 0.573 z-score/σ 28.0 Sigma difference (zN-zN+1)/σ 2.98 PDB Download - Fit 3
-
Rank 3 Cross Correlation Score 0.432 Fisher z-score 0.463 z-score/σ 22.7 Sigma difference (zN-zN+1)/σ 8.35 PDB Download - Fit 4
-
Rank 4 Cross Correlation Score 0.301 Fisher z-score 0.311 z-score/σ 15.2 Sigma difference (zN-zN+1)/σ 15.79 PDB Download - Fit 5
-
Rank 5 Cross Correlation Score 0.298 Fisher z-score 0.307 z-score/σ 15.1 Sigma difference (zN-zN+1)/σ 15.97 PDB Download - Fit 6
-
Rank 6 Cross Correlation Score 0.298 Fisher z-score 0.307 z-score/σ 15.0 Sigma difference (zN-zN+1)/σ 15.99 PDB Download - Fit 7
-
Rank 7 Cross Correlation Score 0.293 Fisher z-score 0.302 z-score/σ 14.8 Sigma difference (zN-zN+1)/σ 16.24 PDB Download - Fit 8
-
Rank 8 Cross Correlation Score 0.290 Fisher z-score 0.298 z-score/σ 14.6 Sigma difference (zN-zN+1)/σ 16.42 PDB Download - Fit 9
-
Rank 9 Cross Correlation Score 0.288 Fisher z-score 0.297 z-score/σ 14.5 Sigma difference (zN-zN+1)/σ 16.49 PDB Download - Fit 10
-
Rank 10 Cross Correlation Score 0.288 Fisher z-score 0.296 z-score/σ 14.5 Sigma difference (zN-zN+1)/σ 16.53 PDB Download