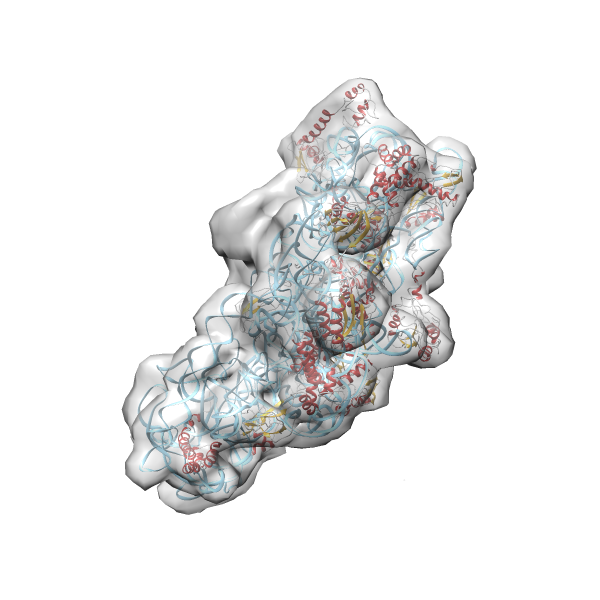
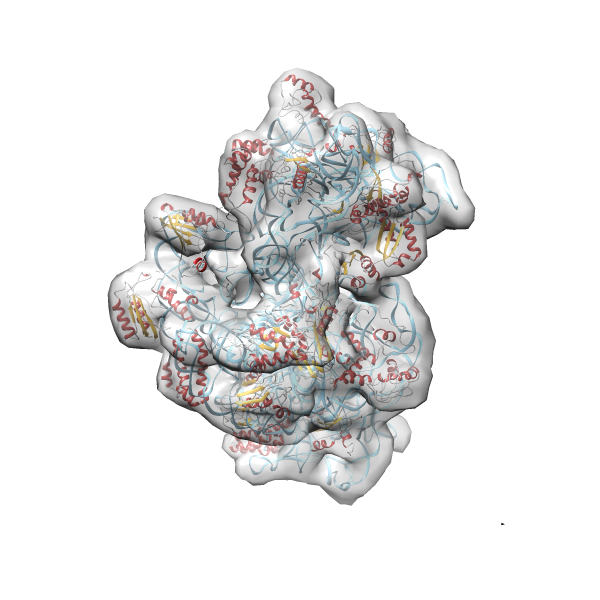
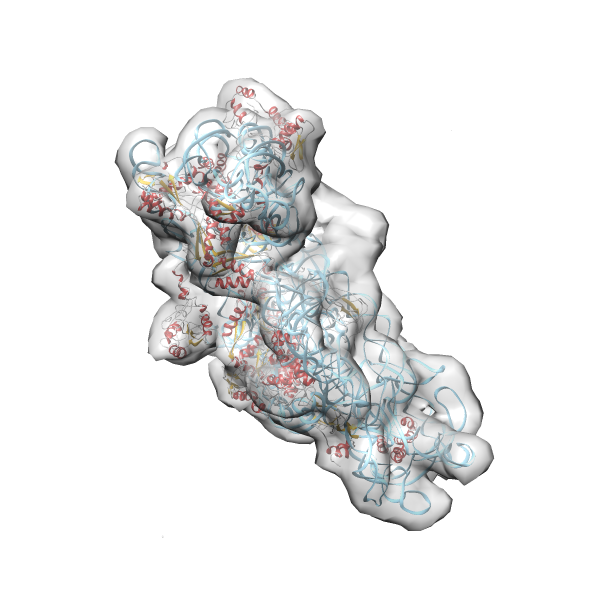
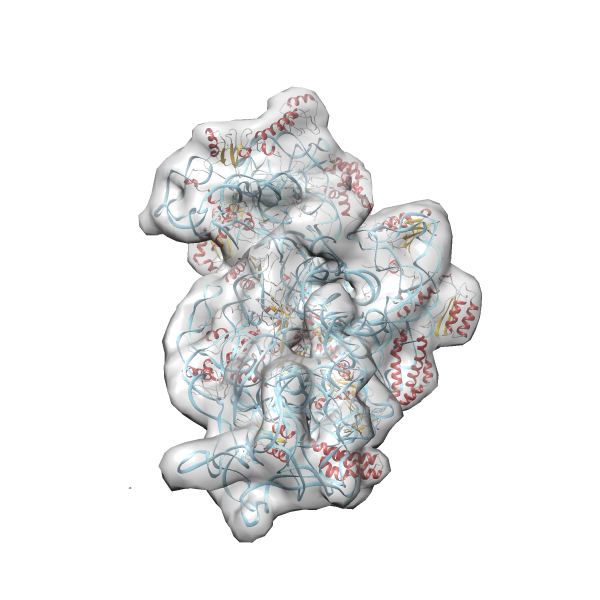
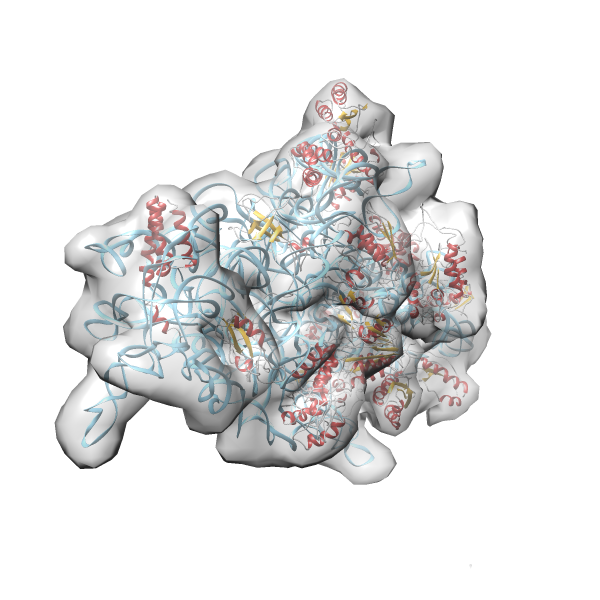
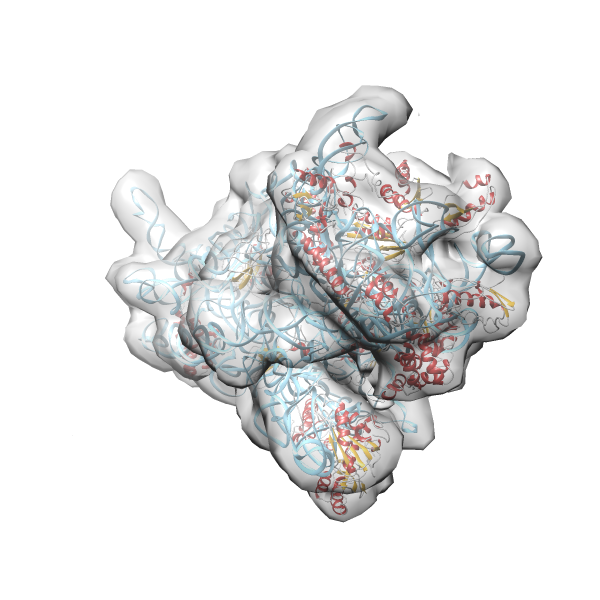
- GTPase RsgA-30S ribosomal subunit-GMPPNP complex
- In this example the 30S ribosomal subunit-GMPPNP complex (PDB entry 2ykr) without the GTPase RsgA (chain W) was fitted into the cryo-EM density map of the full RsgA-30S ribosomal subunit-GMPPNP complex (EMDB entry 1884 - 9.8 Å resolution) with the default server parameters. For this example Powerfit only reports one acceptable solution fitting well into the provided density map.
- Run 2ykr without RsgA - EMD-1884 (Example2)
- Status: FINISHED
- Your PowerFit run has successfully completed.
-
Archive of the complete run: Example2.tgz Archive of all autogenerated images: Example2_images.tgz - Please cite the following papers in your work:
-
G.C.P. van Zundert, M. Trellet, J. Schaarschmidt, Z. Kurkcuoglu, M. David, M. Verlato, A. Rosato and A.M.J.J. Bonvin.
The DisVis and PowerFit web servers: Explorative and Integrative Modeling of Biomolecular Complexes. J. Mol. Biol., Advanced Online Publication (2016).
G.C.P. van Zundert and A.M.J.J. Bonvin (2015)
Fast and sensitive rigid-body fitting into cryo-EM density maps with PowerFit.
AIMS Biophysics 2, 73-87. - and add the following acknowledgment:
- The FP7 WeNMR (project# 261572), H2020 West-Life (project# 675858), EOSC-hub (project# 777536) and the EGI-ACE (project# 101017567) European e-Infrastructure projects are acknowledged for the use of their web portals, which make use of the EGI infrastructure with the dedicated support of CESNET-MCC, INFN-LNL-2, NCG-INGRID-PT, TW-NCHC, CESGA, IFCA-LCG2, UA-BITP, TR-FC1-ULAKBIM, CSTCLOUD-EGI, IN2P3-CPPM, CIRMMP, SURFsara and NIKHEF, and the additional support of the national GRID Initiatives of Belgium, France, Italy, Germany, the Netherlands, Poland, Portugal, Spain, UK, Taiwan and the US Open Science Grid.
- Note: Your results will be stored for 14 days before being removed from the server, please make a backup as soon as possible.
- How would you rate your experience with our portal? sentiment_very_dissatisfied sentiment_dissatisfied sentiment_neutral sentiment_satisfied sentiment_very_satisfied Thank you!done
- Questions / feedback ? ask.bioexcel.eu
- Please also consider giving us some feedback by filling our online survey.
- Solutions
- The table below lists the 15 best non-redundant solutions found by correlation score. The first column shows the rank, column 2 the correlation score, column 3 the Fisher z-score column 4 the zscore as factor of standard deviations (z/σ), and column 5 the sigma difference to the best fit. (see N. Volkmann 2009, and Van Zundert and Bonvin 2016).
-
Rank (N) Cross Correlation Score Fisher z-score z-score/σ Sigma difference
(z1-zN)/σ1 0.666 0.804 183.8 0.00 - Images were generated with UCSF Chimera.
- Fit 1
-
Rank 1 Cross Correlation Score 0.666 Fisher z-score 0.804 z-score/σ 183.8 Sigma difference (zN-zN+1)/σ 0.00 PDB Download